Random Forest Feature Selection
Random Forest feature selection, why we need feature selection?
When we have too many features or variables in the datasets and we want to develop a prediction model like a neural network will take a lot of computational time and more features reduce the accuracy of the prediction model. In such cases, we need to make use of the feature selection Boruta algorithm and which is based on a random forest.
How Boruta works?
Suppose if we have 100 variables in the dataset, each attributes creates shadow attributes, and in each shadow attribute, all the values are shuffled and creates randomness in the dataset.
Based on these datasets will create a classification model with shadow attributes and original attributes and then assess the importance of the attributes.
Random Forest Classification Model
Load Libraries
library(Boruta) library(mlbench) library(caret) library(randomForest)
Getting Data
data("Sonar")
str(Sonar)The dataset contains 208 observations with 61 variables.
'data.frame': 208 obs. of 61 variables: $ V1 : num 0.02 0.0453 0.0262 0.01 0.0762 0.0286 0.0317 0.0519 0.0223 0.0164 ... $ V2 : num 0.0371 0.0523 0.0582 0.0171 0.0666 0.0453 0.0956 0.0548 0.0375 0.0173 ... $ V3 : num 0.0428 0.0843 0.1099 0.0623 0.0481 ... $ V4 : num 0.0207 0.0689 0.1083 0.0205 0.0394 ... $ V5 : num 0.0954 0.1183 0.0974 0.0205 0.059 ... $ V6 : num 0.0986 0.2583 0.228 0.0368 0.0649 ... ............................................... $ V20 : num 0.48 0.782 0.862 0.397 0.464 ... $ V59 : num 0.009 0.0052 0.0095 0.004 0.0107 0.0051 0.0036 0.0048 0.0059 0.0056 ... $ V60 : num 0.0032 0.0044 0.0078 0.0117 0.0094 0.0062 0.0103 0.0053 0.0022 0.004 ... $ Class: Factor w/ 2 levels "M","R": 2 2 2 2 2 2 2 2 2 2 ...
Class is the dependent variable with 2 level factors Mine and Rock.
Feature Selection
set.seed(111) boruta <- Boruta(Class ~ ., data = Sonar, doTrace = 2, maxRuns = 500) print(boruta)
Boruta performed 499 iterations in 1.3 mins.
33 attributes confirmed important: V1, V10, V11, V12, V13 and 28 more; 20 attributes confirmed unimportant: V14, V24, V25, V29, V3 and 15 more; 7 tentative attributes left: V2, V30, V32, V34, V39 and 2 more;
Based on Boruta algorithm 33 attributes are important, 20 attributes are unimportant and 7 are tentative attributes.
plot(boruta, las = 2, cex.axis = 0.7)
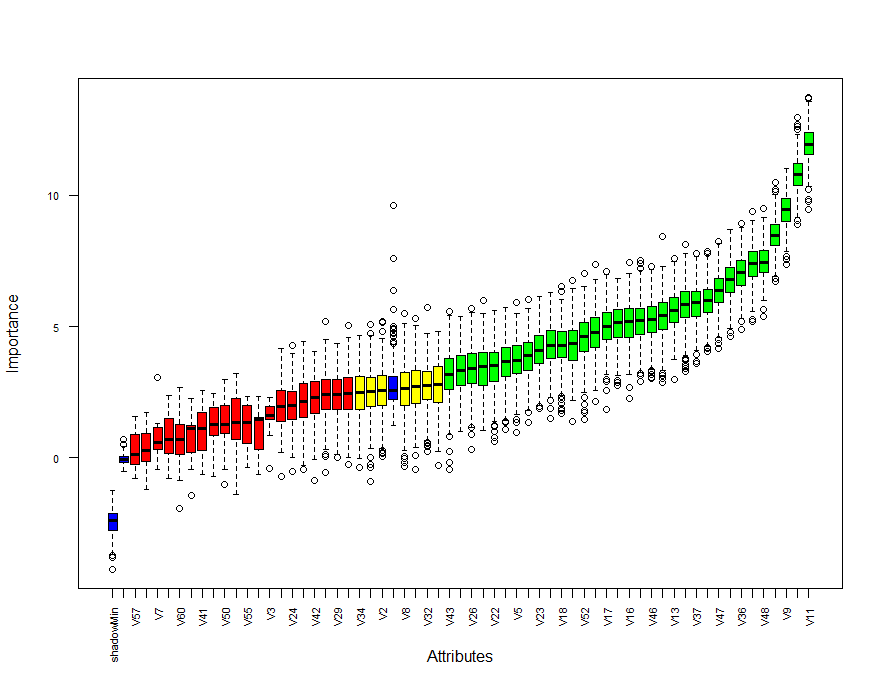
Blue box corresponds to shadow attributes, green color indicates important attributes, yellow boxes are tentative attributes and red boxes are unimportant.
plotImpHistory(boruta)

Tentative Fix
bor <- TentativeRoughFix(boruta) print(bor)
Basically, TentativeRoughFix will take care of tentative attributes that are classified into really important or unimportant classes.
Boruta performed 499 iterations in 1.3 mins.
Tentatives rough fixed over the last 499 iterations. 35 attributes confirmed important: V1, V10, V11, V12, V13 and 30 more; 25 attributes confirmed unimportant: V14, V2, V24, V25, V29 and 20 more;
attStats(boruta)
This will provide complete picture of all the variables.
meanImp medianImp minImp maxImp normHits decision V1 3.63 3.66 1.0746 5.7 0.804 Confirmed V2 2.54 2.55 0.0356 5.2 0.479 Tentative V3 1.52 1.62 -0.4086 2.3 0.000 Rejected V4 5.39 5.43 2.8836 8.4 0.990 Confirmed V5 3.70 3.70 0.9761 5.9 0.814 Confirmed V6 2.12 2.16 -0.4508 4.4 0.090 Rejected V7 0.72 0.59 -0.4309 3.1 0.002 Rejected V8 2.63 2.62 -0.3495 5.5 0.463 Tentative ..................................................... V59 2.40 2.40 -0.5639 5.2 0.200 Rejected V60 0.72 0.69 -1.9414 2.7 0.002 Rejected
Data Partition
Let’s partition the dataset into training datasets and test datasets. Now we want to build the Boruta algorithm for higher model accuracy.
set.seed(222) ind <- sample(2, nrow(Sonar), replace = T, prob = c(0.6, 0.4)) train <- Sonar[ind==1,] test <- Sonar[ind==2,]
Training dataset contains 117 observations and test data set contains 91 observations.
Random Forest Model
set.seed(333) rf60 <- randomForest(Class~., data = train)
Random forest model based on all the varaibles in the dataset
Call: randomForest(formula = Class ~ ., data = train) Type of random forest: classification Number of trees: 500 No. of variables tried at each split: 7 OOB estimate of error rate: 23% Confusion matrix: M R class.error M 51 10 0.16 R 17 39 0.30
OOB error rate is 23%
Prediction & Confusion Matrix – Test
p <- predict(rf60, test) confusionMatrix(p, test$Class)
Confusion Matrix and Statistics
Reference Prediction M R M 46 17 R 4 24 Accuracy : 0.769 95% CI : (0.669, 0.851) No Information Rate : 0.549 P-Value [Acc > NIR] : 1.13e-05 Kappa : 0.52 Mcnemar's Test P-Value : 0.00883 Sensitivity : 0.920 Specificity : 0.585 Pos Pred Value : 0.730 \ Neg Pred Value : 0.857 Prevalence : 0.549 Detection Rate : 0.505 Detection Prevalence : 0.692 Balanced Accuracy : 0.753 'Positive' Class : M
Based on this model accuracy is around 76%. Now let’s make use of the Boruta model.
getNonRejectedFormula(boruta)
rfboruta <- randomForest(Class ~ V1 + V2 + V4 + V5 + V8 + V9 + V10 + V11 + V12 + V13 +
V15 + V16 + V17 + V18 + V19 + V20 + V21 + V22 + V23 + V26 +
V27 + V28 + V30 + V31 + V32 + V34 + V35 + V36 + V37 + V39 +
V43 + V44 + V45 + V46 + V47 + V48 + V49 + V51 + V52 + V54, data = train)Call: randomForest(formula = Class ~ V1 + V2 + V4 + V5 + V8 + V9 + V10 + V11 + V12 + V13 + V15 + V16 + V17 + V18 + V19 + V20 + V21 + V22 + V23 + V26 + V27 + V28 + V30 + V31 + V32 + V34 + V35 + V36 + V37 + V39 + V43 + V44 + V45 + V46 + V47 + V48 + V49 + V51 + V52 + V54, data = train) Type of random forest: classification Number of trees: 500 No. of variables tried at each split: 6 OOB estimate of error rate: 22% Confusion matrix: M R class.error M 52 9 0.15 R 17 39 0.30
Now you can see that the OOB error rate reduced from 23% to 22%.
p <- predict(rfboruta, test) confusionMatrix(p, test$Class)
Confusion Matrix and Statistics
Linear Discriminant Analysis in R
Reference Prediction M R M 45 15 R 5 26 Accuracy : 0.78 95% CI : (0.681, 0.86) No Information Rate : 0.549 P-Value [Acc > NIR] : 3.96e-06 Kappa : 0.546 Mcnemar's Test P-Value : 0.0442 Sensitivity : 0.900 Specificity : 0.634 Pos Pred Value : 0.750 Neg Pred Value : 0.839 Prevalence : 0.549 Detection Rate : 0.495 Detection Prevalence : 0.659 Balanced Accuracy : 0.767 'Positive' Class : M
Accuracy increased from 76% to 78%.
getConfirmedFormula(boruta)
rfconfirm <- randomForest(Class ~ V1 + V4 + V5 + V9 + V10 + V11 + V12 + V13 + V15 + V16 + V17 + V18 + V19 + V20 + V21 + V22 + V23 + V26 + V27 + V28 + V31 + V35 + V36 + V37 + V43 + V44 + V45 + V46 + V47 + V48 + V49 + V51 + V52, data = train)
Call:
randomForest(formula = Class ~ V1 + V4 + V5 + V9 + V10 + V11 + V12 + V13 + V15 + V16 + V17 + V18 + V19 + V20 + V21 + V22 + V23 + V26 + V27 + V28 + V31 + V35 + V36 + V37 + V43 + V44 + V45 + V46 + V47 + V48 + V49 + V51 + V52, data = train) Type of random forest: classification Number of trees: 500 No. of variables tried at each split: 5 OOB estimate of error rate: 20% Confusion matrix: M R class.error M 53 8 0.13 R 15 41 0.27
Now you can see that based on important attributes is OOB error rate is further reduced into 20%.
Conclusion
Based on feature selection you can increase the accuracy of the model and if you are using neural network types of model can reduce the computational time also.
KNN Algorithm Machine Learning



Could you please check variable “rfboura” . I get error and I don’t see it generated anywhere in code. rf60 works but don’t get ,78 accuracy. “boruta” give me 2nd error code. Everything else works up to that section.
Thank you
p <- predict(rfboruta, test)
confusionMatrix(p, test$Class)
Errorcode1: Error in predict(rfboruta, test) : object 'rfboruta' not found
Errorcode2: Error in UseMethod("predict") :
no applicable method for 'predict' applied to an object of class "Boruta"
rfboruta based on getNonRejectedFormula ..updated the code now.
Thanks
Thanks