Linear Discriminant Analysis in R
Linear Discriminant Analysis in R, originally developed by R A Fisher in 1936 to classify subjects into one of the two clearly defined groups.
It was later expanded to classify subjects into more than two groups.
Linear Discriminant Analysis (LDA) is a dimensionality reduction technique. LDA used for dimensionality reduction to reduce the number of dimensions (i.e. variables) in a dataset while retaining as much information as possible.
Basically, it helps to find the linear combination of original variables that provide the best possible separation between the groups.
Customer Segmentation analysis in R
The basic purpose is to estimate the relationship between a single categorical dependent variable and a set of quantitative independent variables.
The major applications or examples are
- Predicting success or failure of new products
- Accepting or rejecting admission to an applicant.
- Predicting credit risk category for a person
- Classifying patients into different categories.
Let’s look into the iris data set for further analysis.
Load Library
library(klaR) library(psych) library(MASS) library(ggord) library(devtools)
Getting Data
data("iris")
str(iris)Total 150 observations and 5 variables contains in the iris dataset.
'data.frame': 150 obs. of 5 variables: $ Sepal.Length: num 5.1 4.9 4.7 4.6 5 5.4 4.6 5 4.4 4.9 ... $ Sepal.Width : num 3.5 3 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1 ... $ Petal.Length: num 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1.5 ... $ Petal.Width : num 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 ... $ Species : Factor w/ 3 levels "setosa","versicolor",..: 1 1 1 1 1 1 1 1 1 1 ...
First will create a scatterplot for the first four numerical variables. The gap between the points given is zero.
pairs.panels(iris[1:4],
gap = 0,
bg = c("red", "green", "blue")[iris$Species],
pch = 21)
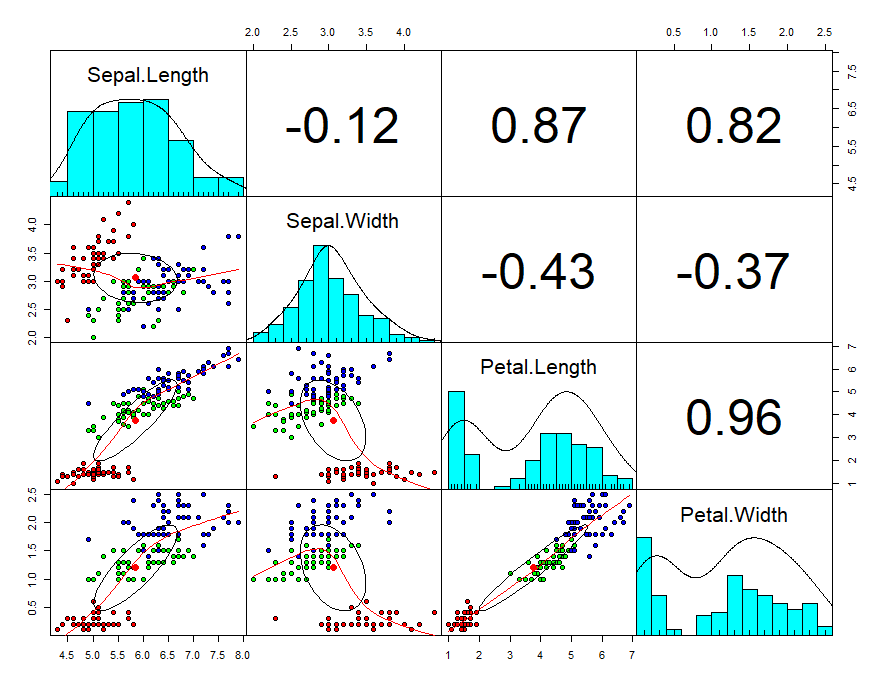
Now you can see in the plot, scatter diagram, histogram, and correlation values.
Now we want to create the best separation groups based on these species.
Data partition
Let’s create a training dataset and test dataset for prediction and testing purposes. 60% dataset used for training purposes and 40$ used for testing purposes.
set.seed(123) ind <- sample(2, nrow(iris), replace = TRUE, prob = c(0.6, 0.4)) training <- iris[ind==1,] testing <- iris[ind==2,]
Linear discriminant analysis
linear <- lda(Species~., training) linear
Call:
lda(Species ~ ., data = training) Prior probabilities of groups: setosa versicolor virginica 0.3837209 0.3139535 0.3023256 Group means: Sepal.Length Sepal.Width Petal.Length Petal.Width setosa 4.975758 3.357576 1.472727 0.2454545 versicolor 5.974074 2.751852 4.281481 1.3407407 virginica 6.580769 2.946154 5.553846 1.9807692 Coefficients of linear discriminants: LD1 LD2 Sepal.Length 1.252207 -0.1229923 Sepal.Width 1.115823 2.2711963 Petal.Length -2.616277 -0.7924520 Petal.Width -2.156489 2.6956343 The proportion of trace: LD1 LD2 0.9937 0.0063
Based on the training dataset, 38% belongs to setosa group, 31% belongs to versicolor groups and 30% belongs to virginica groups.
The first discriminant function is a linear combination of the four variables.
Percentage separations achieved by the first discriminant function is 99.37% and second is 0.63%
attributes(linear)
[1] "prior" "counts" "means" "scaling" "lev" "svd" "N" "call" "terms" [10] "xlevels" $class [1] "lda"
Histogram
Stacked histogram for discriminant function values.
p <- predict(linear, training) ldahist(data = p$x[,1], g = training$Species)
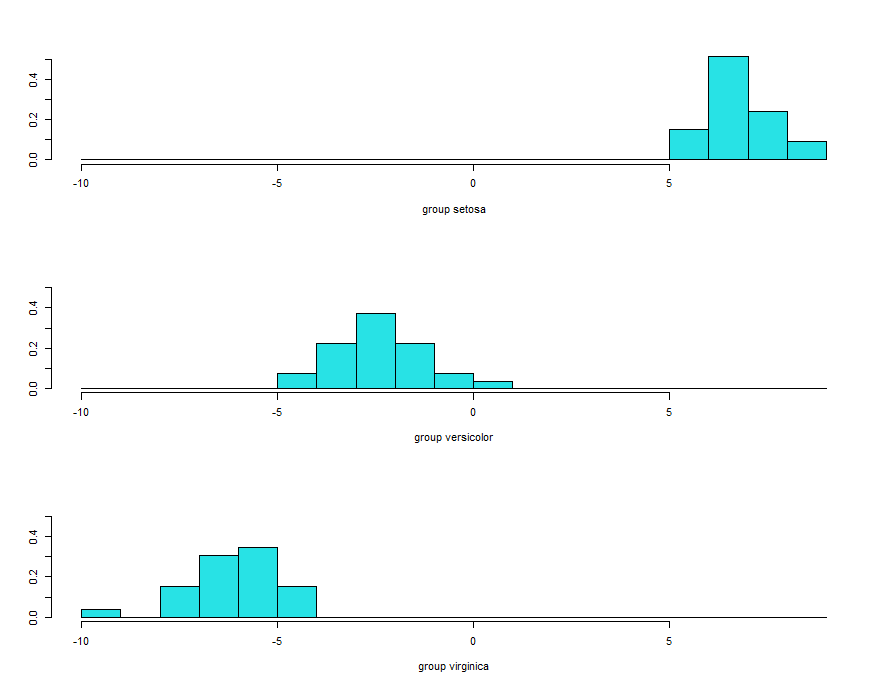
These histograms are based on ld1. It’s clearly evident that no overlaps between first and second and first and third species. But some overlap observed between the second and third species.
ldahist(data = p$x[,2], g = training$Species)
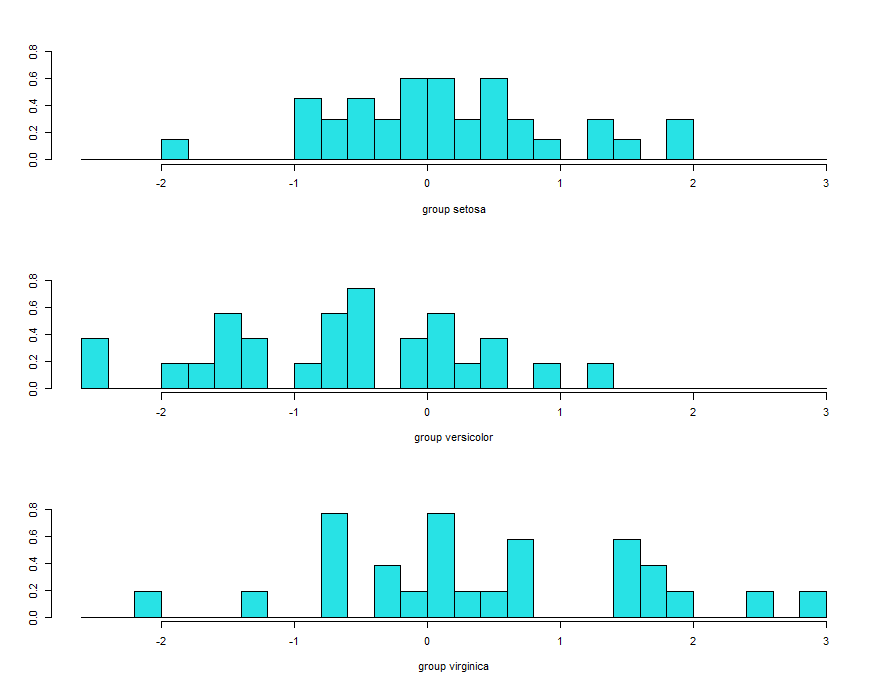
histogram based on lda2 showing complete overlap and its not good.
Bi-Plot
ggord(linear, training$Species, ylim = c(-10, 10))
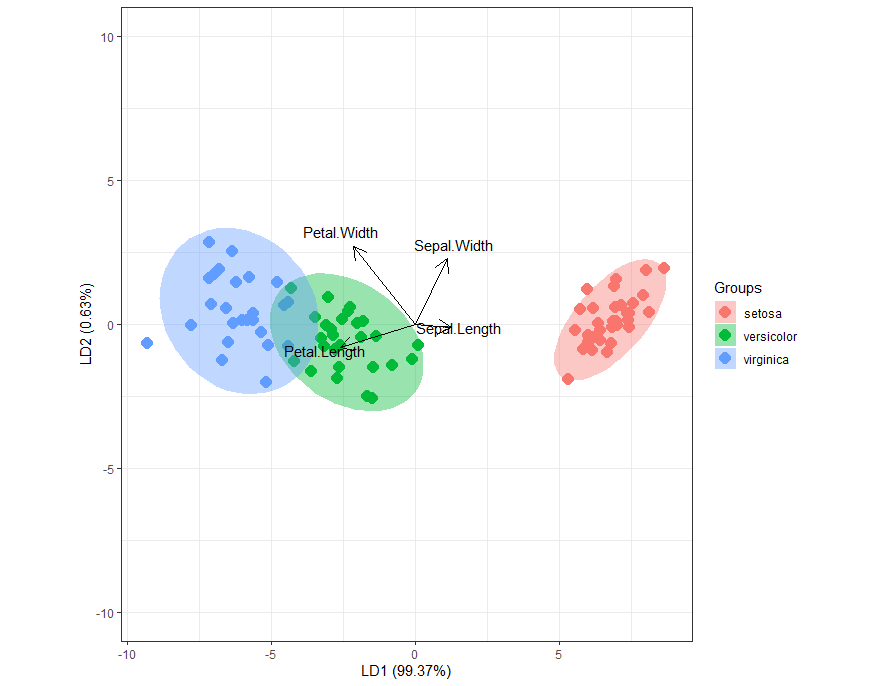
Biplot based on LD1 and LD2. Setosa separated very clearly and some overlap observed between Versicolor and virginica.
Based on arrows, Sepal width and sepal length explained more for setosa, petal width and petal length explained more for versicolor and virginica.
Partition plot
It provides the classification of each and every combination in the training dataset.
partimat(Species~., data = training, method = "lda")
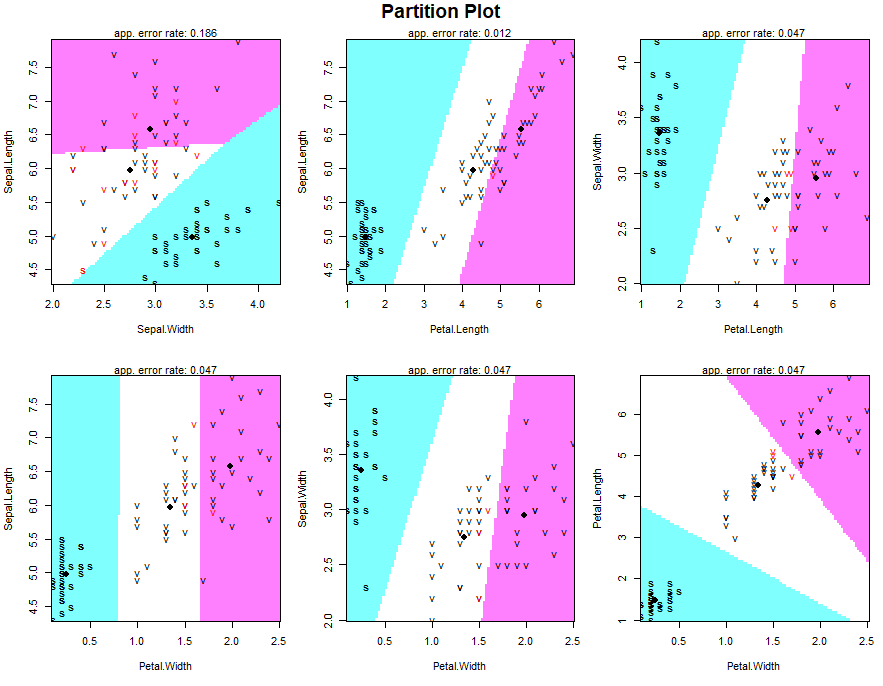
partimat(Species~., data = training, method = "qda")

Confusion matrix and accuracy – training data
p1 <- predict(linear, training)$class tab <- table(Predicted = p1, Actual = training$Species) tab
Let’s see the correct classifications and miss classifications.
Repeated Measures of ANOVA in R
Actual Predicted setosa versicolor virginica setosa 33 0 0 versicolor 0 26 1 virginica 0 1 25
In the training dataset total correct classification is 33+26+25=84
sum(diag(tab))/sum(tab)
The accuracy of the model is around 0.9767442
Confusion matrix and accuracy – testing data
p2 <- predict(linear, testing)$class tab1 <- table(Predicted = p2, Actual = testing$Species) tab1
Actual Predicted setosa versicolor virginica setosa 17 0 0 versicolor 0 22 0 virginica 0 1 24
sum(diag(tab1))/sum(tab1)
The accuracy of the model is around .984375
Conclusion
Histogram and Biplot provide useful insights and helpful for interpretations and if there is not a great difference in the group covariance matrices, then the linear discriminant analysis will perform as well as quadratic. LDA is not useful for solving non-linear problems.
KNN Algorithm Machine Learning






Thanks for a great post and for working with data that can be accessed to follow along.
Thanks a lot…
The plot suggestions really helped – thank you!
Thanks for your reply…