How to Perform Tukey HSD Test in R
Tukey HSD Test in R, When there are three or more independent groups, we apply a one-way ANOVA to see if there is a significant difference.
The p-value for one-way ANOVA is less than 0.05 indicate that at least one of the treatment groups differs from the others.
One way ANOVA tells us whether the means of the groups are significantly different or not, but we still need to do a post hoc multiple comparison test to dig further.
One of the widely used methods is the Tukey HSD test.
How to do data reshape in R? » Data Reshaping »
What is special with Tukey HSD?
It is a multiple range test similar to the LSD test except that Tukey utilized the honestly significant difference (HSD) test or the w-procedure.
Any two treatments that mean having a difference more than honestly significant difference are said to be significantly different, otherwise not.
Tukey HSD Test in R
The Tukey HSD test allows for all possible pairwise comparisons while keeping the family-wise error rate low.
Step 1: ANOVA Model
For the difference identification, establish a data frame with three independent groups and fit a one-way ANOVA model.
set.seed(1045)
data <- data.frame(group = rep(c("P1", "P2", "P3"), each = 40),
values = c(rnorm(40, 0, 3),rnorm (40, 0, 6),rnorm (40, 1, 5)))
head(data)one-way ANOVA model
model <- aov(values~group, data=data) summary(model)
Df Sum Sq Mean Sq F value Pr(>F) group 2 400.7 200.35 9.9 0.000107 *** Residuals 117 2367.8 20.24 --- Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’
The observed p-value from the ANOVA table is less than 0.05, indicating that there is enough evidence to conclude that the group means are not equal.
How to read a stock chart pattern Chart Reading Technique
Step 2: Perform Tukey HSD Test.
TukeyHSD(model, conf.level=.95)
Tukey multiple comparisons of means 95% family-wise confidence level Fit: aov(formula = values ~ group, data = data) $group diff lwr upr p adj P2-P1 -1.524566 -3.9125188 0.863387 0.2873372 P3-P1 2.882295 0.4943425 5.270248 0.0135957 P3-P2 4.406861 2.0189084 6.794814 0.0000765
P3 vs P1 and P3 vs P2 are significantly different at the 95 percent confidence level, according to the Tukey HSD test.
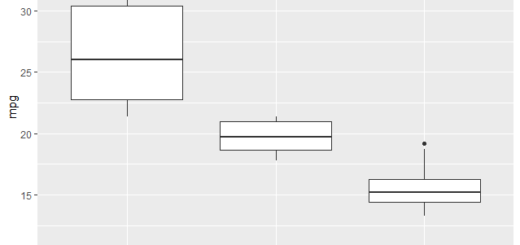
Step 3: Visualization
TukeyHSD() function allows us to visualize the confidence intervals
plot(TukeyHSD(model, conf.level=.95), las = 2)

Correlation Analysis in R? » Karl Pearson correlation coefficient »


Dear finnstats,
I use aov() with weighted means, then I use TukeyHSD() and it shows the following message:
Error in result[, i] <- prj[, select, drop = FALSE] %*% rep.int(1, df[i]) :
number of items to replace is not a multiple of replacement length
I'd like to know if you could help me fix this error.
Thanks a lot