Two-Way ANOVA Example in R-Quick Guide
Two-Way ANOVA Example in R, the two-way ANOVA test is used to compare the effects of two grouping variables (A and B) on a response variable at the same time.
Factors are another name for grouping variables. Levels are the several categories (groups) of a component. The number of levels varies depending on the element.
The balanced design occurs when the sample sizes within cells are equal. The conventional two-way ANOVA test can be used in this circumstance.
The ANOVA test should be handled differently when the sample sizes within each level of the independent variables are not the same (instance of unbalanced designs).
This article shows how to use R software to compute two-way ANOVA tests for balanced and unbalanced designs.
Hypotheses two-way ANOVA.
There is no difference in factor A’s means.
There is no difference in factor B’s means.
Factors A and B do not interact in any way.
For examples 1 and 2, the alternative hypothesis is that the means are not equal.
The alternate hypothesis for scenario 3 is that A and B have an interaction.
Two-way ANOVA test assumptions
Like all ANOVA tests, two-way ANOVA assumes that the observations within each cell are normally distributed with equal variances.
After fitting ANOVA, we’ll teach you how to double-check these assumptions.
In R, create a two-way ANOVA test using balanced designs
When we have equal sample sizes within levels of our independent grouping levels, we call it a balanced design.
We’ll use the built-in R data set ToothGrowth for this. It includes information from a study on the effects of vitamin C on tooth growth in Guinea pigs.
The trial used 60 pigs who were given one of three vitamin C dose levels (0.5, 1, or 2 mg/day) via one of two administration routes (orange juice or ascorbic acid) (a form of vitamin C and coded as VC).
A sample of the data is presented below, which includes tooth length measurements.
data <- ToothGrowth
We use the function sample n() [in the dplyr package] to display a random sample of the data to get an idea of how the data looks. Install dplyr first if you don’t already have it.
install.packages("dplyr")Show a random sample
set.seed(123) dplyr::sample_n(data, 5)
len supp dose 1 15.2 OJ 0.5 2 22.5 VC 1.0 3 25.5 OJ 2.0 4 17.3 VC 1.0 5 7.3 VC 0.5
Check the structure
str(data)
'data.frame': 60 obs. of 3 variables: $ len : num 4.2 11.5 7.3 5.8 6.4 10 11.2 11.2 5.2 7 ... $ supp: Factor w/ 2 levels "OJ","VC": 2 2 2 2 2 2 2 2 2 2 ... $ dose: num 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 ...
R treats “dose” as a numeric variable based on the output. As follows, we’ll transform it to a factor variable (i.e., grouping variable).
Recode the levels after converting the dose to a factor.
data$dose <- factor(data$dose,
levels = c(0.5, 1, 2),
labels = c("D0.5", "D1", "D2"))
str(data)data.frame': 60 obs. of 3 variables: $ len : num 4.2 11.5 7.3 5.8 6.4 10 11.2 11.2 5.2 7 ... $ supp: Factor w/ 2 levels "OJ","VC": 2 2 2 2 2 2 2 2 2 2 ... $ dose: Factor w/ 3 levels "D0.5","D1","D2": 1 1 1 1 1 1 1 1 1 1 ..
Question: Does tooth length vary according to on supp and dose?
Make frequency tables by:
table(data$supp,data$dose)
D0.5 D1 D2 OJ 10 10 10 VC 10 10 10
We have 2 X 3 design cells, each with 10 individuals and the factors supp and dose. We have a well-balanced design here.
Because this is the most basic situation, we’ll show how to analyze data from balanced designs in the next sections.
Visualize your information
To depict group differences, box plots and line plots can be used:
To visualize the data grouped by the levels of the two factors, use a box plot.
The mean (or another summary) of the answer for two-way combinations of factors is plotted in a two-way interaction plot, indicating probable interactions.
Read R base graphs to learn how to utilise them. For an easy ggplot2-based data visualisation, we’ll use the ggpubr R tool.
install.packages("ggpubr")Visualize your data with ggpubr:
Plot tooth length (“len”) by groups (“dose”)
Color box plot by a second group: “supp”
library("ggpubr")
ggboxplot(data, x = "dose", y = "len", color = "supp",
palette = c("#00AFBB", "#E7B800"))
Add error bars: mean_se
library("ggpubr")
ggline(data, x = "dose", y = "len", color = "supp",
add = c("mean_se", "dotplot"),
palette = c("#00AFBB", "#E7B800"))
Type the following scripts if you still want to utilize R basic graphs:
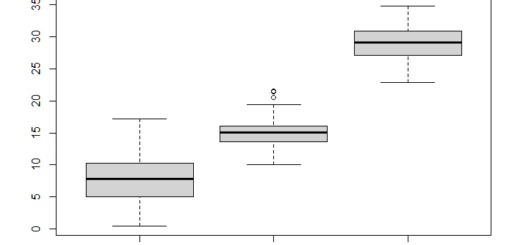
Box plot with two variable factors
boxplot(len ~ supp * dose, data=data, frame = FALSE,
col = c("#00AFBB", "#E7B800"), ylab="Tooth Length")
Two-way interaction plot
interaction.plot(x.factor = data$dose, trace.factor = data$supp,
response = data$len, fun = mean,
type = "b", legend = TRUE,
xlab = "Dose", ylab="Tooth Length",
pch=c(1,19), col = c("#00AFBB", "#E7B800"))
Compute a two-way ANOVA test
We’re curious if tooth length is affected by supp and dose.
This question can be answered using the R function aov(). Summary of the function The analysis of the variance model is summarised using aov().
res.aov2 <- aov(len ~ supp + dose, data = data) summary(res.aov2)
Df Sum Sq Mean Sq F value Pr(>F) supp 1 205.4 205.4 11.45 0.0013 ** dose 1 2224.3 2224.3 123.99 6.31e-16 *** Residuals 57 1022.6 17.9 --- Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1--- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
The columns F value and Pr(>F) in the output corresponding to the p-value of the test.
We may deduce from the ANOVA table that both supp and dose are statistically significant. The most important factor variable is dosage.
These findings led us to anticipate that modifying the delivery technique (supp) or the vitamin C dose will have a major impact on the mean tooth length.
The above-fitted model is not referred to as an additive model. It is presumptively assumed that the two-factor variables are unrelated.
Replace the plus symbol (+) with an asterisk (*) if you think these two variables will interact to create a synergistic effect.
res.aov3 <- aov(len ~ supp * dose, data = data) summary(res.aov3)
Df Sum Sq Mean Sq F value Pr(>F) supp 1 205.4 205.4 12.317 0.000894 *** dose 1 2224.3 2224.3 133.415 < 2e-16 *** supp:dose 1 88.9 88.9 5.333 0.024631 * Residuals 56 933.6 16.7 --- Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
The two primary effects (supplement and dose), as well as their interaction, are statistically significant.
In situations where the interaction is not significant, the additive model should be used.
Interpret the outcomes
Based on the p-values and a significance level of 0.05, you may deduce the following from the ANOVA results:
Supp has a p-value <0.05 (significant), indicating that varied levels of supp are associated with varying tooth lengths.
The dose p-value <0.05 (significant), indicating that differing treatment levels are linked with substantial differences in tooth length.
The interaction between supp*dose has a p-value of 0.02 (significant), indicating that the connection between dose and tooth length is influenced by the supp technique.
Make a few summary statistics.
Using the dplyr R package, compute mean and SD by groups:
require("dplyr")
group_by(data, supp, dose) %>%
summarise(
count = n(),
mean = mean(len, na.rm = TRUE),
sd = sd(len, na.rm = TRUE)
)supp dose count mean sd <fct> <dbl> <int> <dbl> <dbl> 1 OJ 0.5 10 13.2 4.46 2 OJ 1 10 22.7 3.91 3 OJ 2 10 26.1 2.66 4 VC 0.5 10 7.98 2.75 5 VC 1 10 16.8 2.52 6 VC 2 10 26.1 4.80
Multiple pairwise comparisons between group means
A significant p-value in an ANOVA test shows that some of the group means differ, but we don’t know which pairings of groups differ.
Multiple pairwise comparisons can be performed to see if the mean differences between certain pairs of groups are statistically significant.
Multiple Tukey pairwise comparisons
We can compute Tukey HSD (Tukey Honest Significant Differences, R function: TukeyHSD()) for doing numerous pairwise comparisons between the means of groups because the ANOVA test is significant.
The fitted ANOVA is passed to TukeyHD() as an input.
We don’t need to run the test on the “supp” variable because it only has two levels, both of which have been shown to be substantially different using the ANOVA test.
As a result, the Tukey HSD test will only be performed on the factor variable “dosage.”
Best Data Science YouTube Tutorials Free to Learn
TukeyHSD(res.aov3, which = "dose")
Tukey multiple comparisons of means 95% family-wise confidence level Fit: aov(formula = len ~ supp + dose + supp:dose, data = data) $dose diff lwr upr p adj D1-D0.5 9.130 6.362488 11.897512 0.0e+00 D2-D0.5 15.495 12.727488 18.262512 0.0e+00 D2-D1 6.365 3.597488 9.132512 2.7e-06
diff: the difference between the two groups’ means lwr, upr: bottom and upper-end points of the confidence interval at 95 percent (default) p adj: p-value after multiple comparisons adjustment
The output shows that all pairwise comparisons with an adjusted p-value of 0.05 are significant.
Using the multcomp package, perform several comparisons.
The function glht() [in the multcomp package] can be used to do multiple comparison processes for an ANOVA. General linear hypothesis tests are abbreviated as glht. The following is a simplified format:
library(multcomp) summary(glht(res.aov2, linfct = mcp(dose = "Tukey")))
Simultaneous Tests for General Linear Hypotheses
Multiple Comparisons of Means: Tukey Contrasts Fit: aov(formula = len ~ supp + dose, data = my_data)Linear Hypotheses: Estimate Std. Error t value Pr(>|t|) D1 - D0.5 == 0 9.130 1.210 7.543 <1e-05 *** D2 - D0.5 == 0 15.495 1.210 12.802 <1e-05 *** D2 - D1 == 0 6.365 1.210 5.259 <1e-05 *** --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 (Adjusted p values reported -- single-step method)
T-test on pairs
Pairwise comparisons between group levels with various testing corrections can also be calculated using the pairwise.t.test() function.
pairwise.t.test(data$len, data$dose, p.adjust.method = "BH")
Pairwise comparisons using t-tests with pooled SD
data: data$len and data$dose D0.5 D1 D1 1.0e-08 - D2 4.4e-16 1.4e-05 Method for adjusting the P value: BH
The validity of ANOVA assumptions should be checked.
The data must be regularly distributed, and the variation between groups must be homogeneous. With certain diagnostic plots, we can verify this.
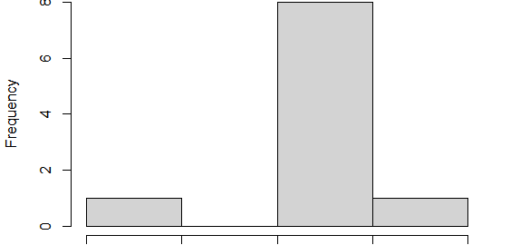
Examine the assumption of homogeneity of variance.
The residuals versus fits graphic are used to assess for variance homogeneity. There are no obvious correlations between residuals and fitted values (the mean of each group) in the plot below, which is good. As a result, we can assume that the variances are homogeneous.
1. Homogeneity of variances
plot(res.aov3, 1)

Two-Way ANOVA Test in R
Outliers are found at points 32 and 23, which can have a significant impact on normalcy and variance homogeneity. To meet the test assumptions, it can be beneficial to remove outliers.
Check the homogeneity of variances with Levene’s test. The leveneTest() method [from the car package] will be used:
library(car) leveneTest(len ~ supp*dose, data = data)
Levene’s Test for Homogeneity of Variance (center = median)
Df F value Pr(>F) group 5 1.7086 0.1484 54
The p-value is not less than the significance level of 0.05, as seen in the output above. This indicates that there is no indication that the variance across groups is statistically significant.
As a result, we can infer that the variations in the different treatment groups are homogeneous.
Examine the assumption of normality.
The residuals’ normality plot. The residuals quantiles are displayed against the normal distribution quantiles in the graph below. Also plotted is a 45-degree reference line.
The residuals’ normal probability plot is used to confirm that the residuals are normally distributed.
The residuals’ normal probability plot should roughly follow a straight line.
2. Normality
plot(res.aov3, 2)

We can infer normalcy because all of the points lie roughly along this reference line.
The Shapiro-Wilk test on the ANOVA residuals (W = 0.98, p = 0.5), which finds no evidence of normality violation, supports the previous conclusion.
Extract the residuals
aov_residuals <- residuals(object = res.aov3)
Run Shapiro-Wilk test
shapiro.test(x = aov_residuals )
Shapiro-Wilk normality test
data: aov_residuals W = 0.98499, p-value = 0.6694
In R, create a two-way ANOVA test for unbalanced designs.
In an unbalanced design, each group has an unequal number of participants.
In an imbalanced design, there are three essentially distinct approaches to do an ANOVA. Type-I, Type-II, and Type-III sums of squares are the three types.
To keep things simple, the Type-III sums of squares are the recommended method.
When the design is balanced, all three ways produce the same result. When the design is unbalanced, however, they do not produce the same outcomes.
For unbalanced designs, the function Anova() [in the car package] can be used to compute a two-way ANOVA test.
Install the software on your PC first. Type install.packages(“car”) in R. Then:
library(car) my_anova <- aov(len ~ supp * dose, data = data)
Anova(my_anova, type = "III") Anova Table (Type III tests) Response: len Sum Sq Df F value Pr(>F) (Intercept) 1750.33 1 132.730 3.603e-16 *** supp 137.81 1 10.450 0.002092 ** dose 885.26 2 33.565 3.363e-10 *** supp:dose 108.32 2 4.107 0.021860 * Residuals 712.11 54 --- Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
That’s all from this post…Keep in touch with us.